§2.2.1Replicationof prokaryotes
Initiation(起始)
1.Recognize the startsite(识别复制起始位点)
2.Unwind dsDNA(解链)
3. Primer synthesis(合成引物)
Startsite
★Thereplication starts at a particular point called origin(复制起始点).
★Theorigin of E. coli, ori C, is at the location of 82.
★Thestructure of the origin is 248 bp long and AT-rich.
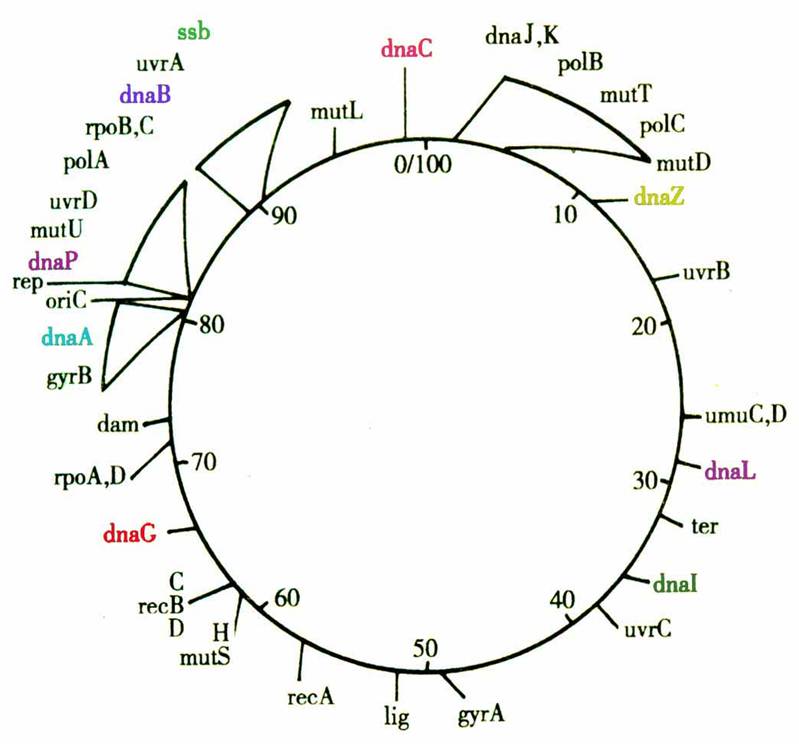
Structureof ori C in E.coli 
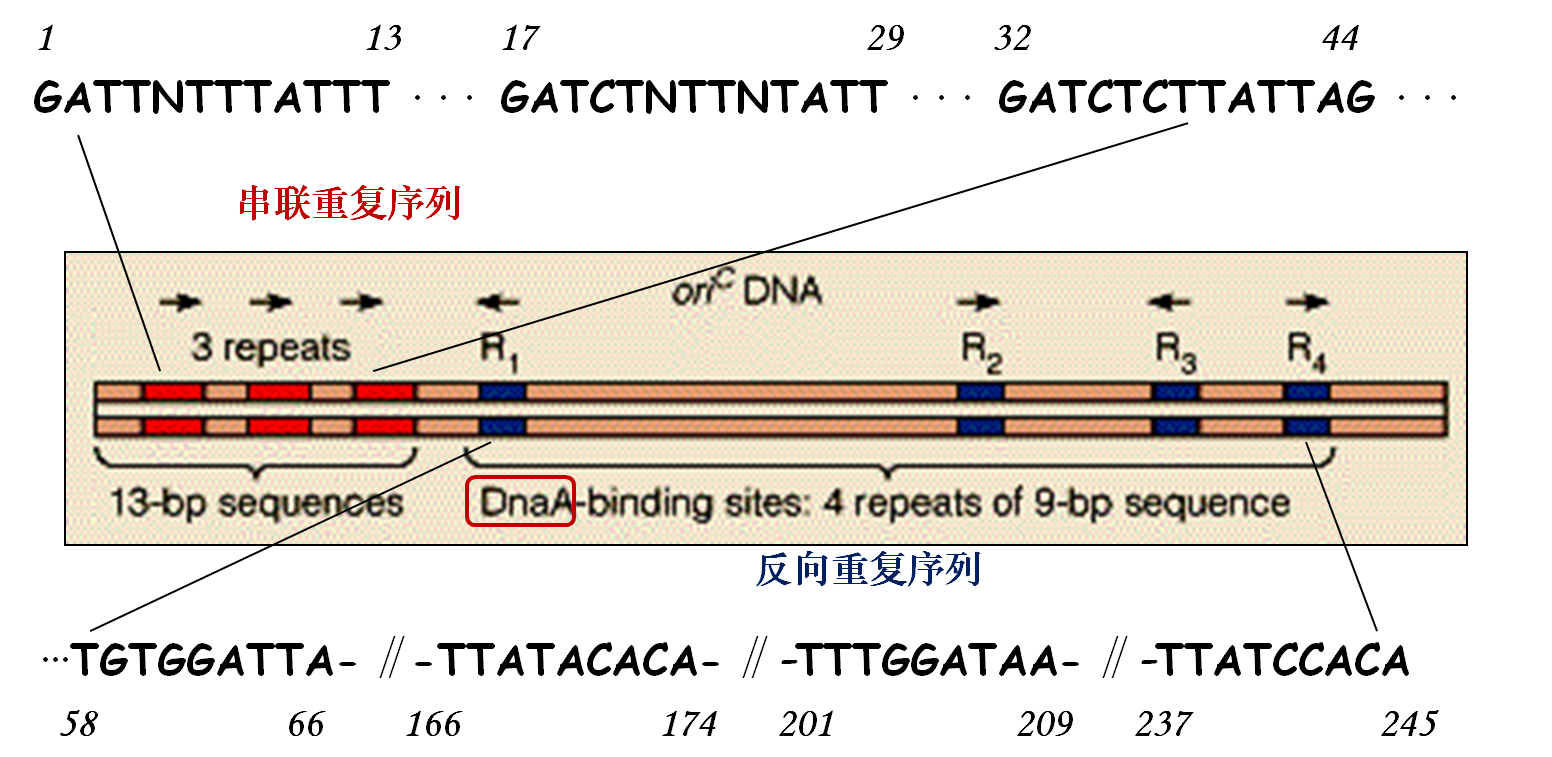
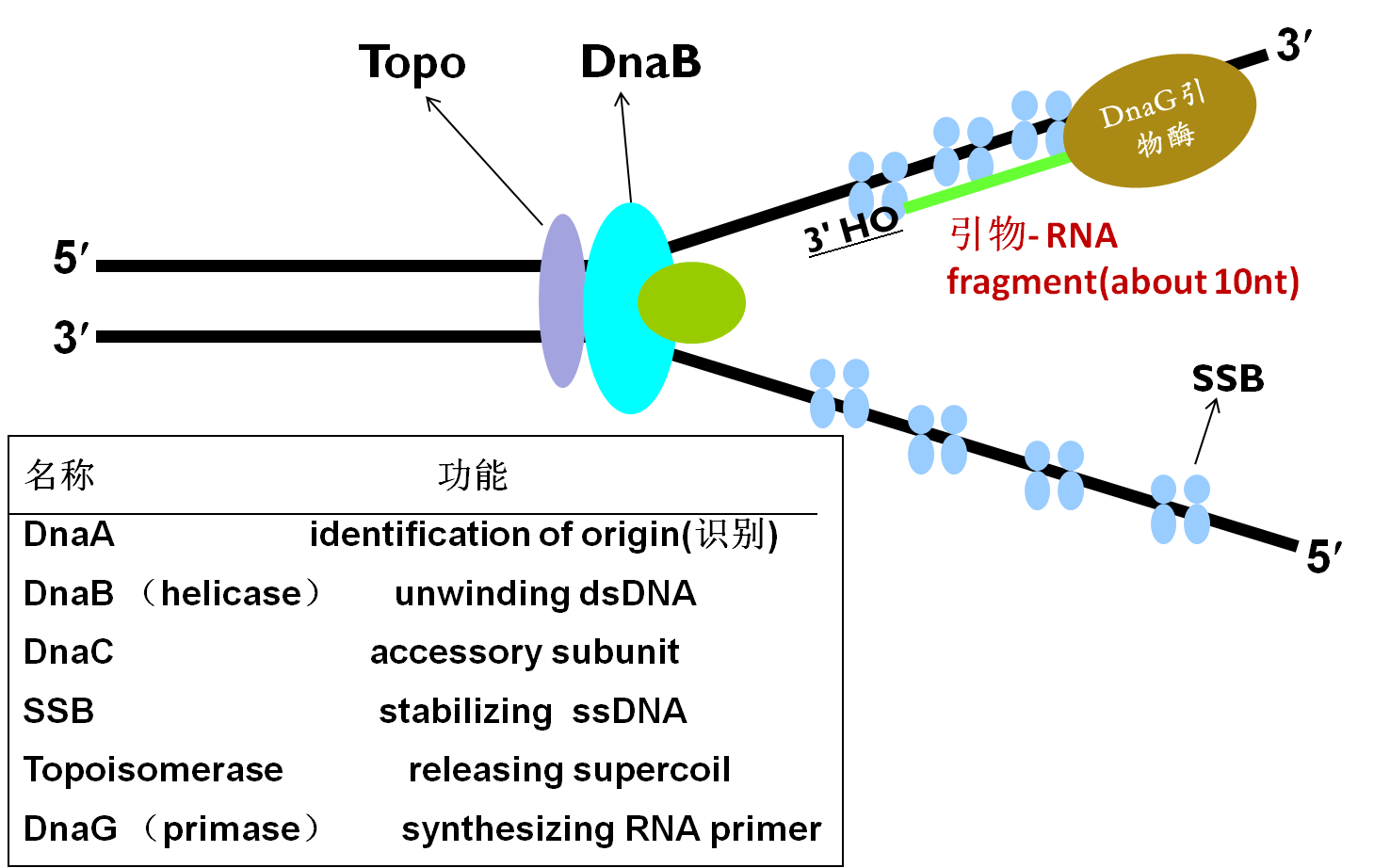
Initiation process
★DnaA recognizes ori C.
★DnaB and DnaC open the local AT-rich region, and move downstream further to separate enough space.
★DnaA is replaced gradually.
★SSB protein binds the complex to stabilize ssDNA.
Ready for elongation
Elongation(延长)

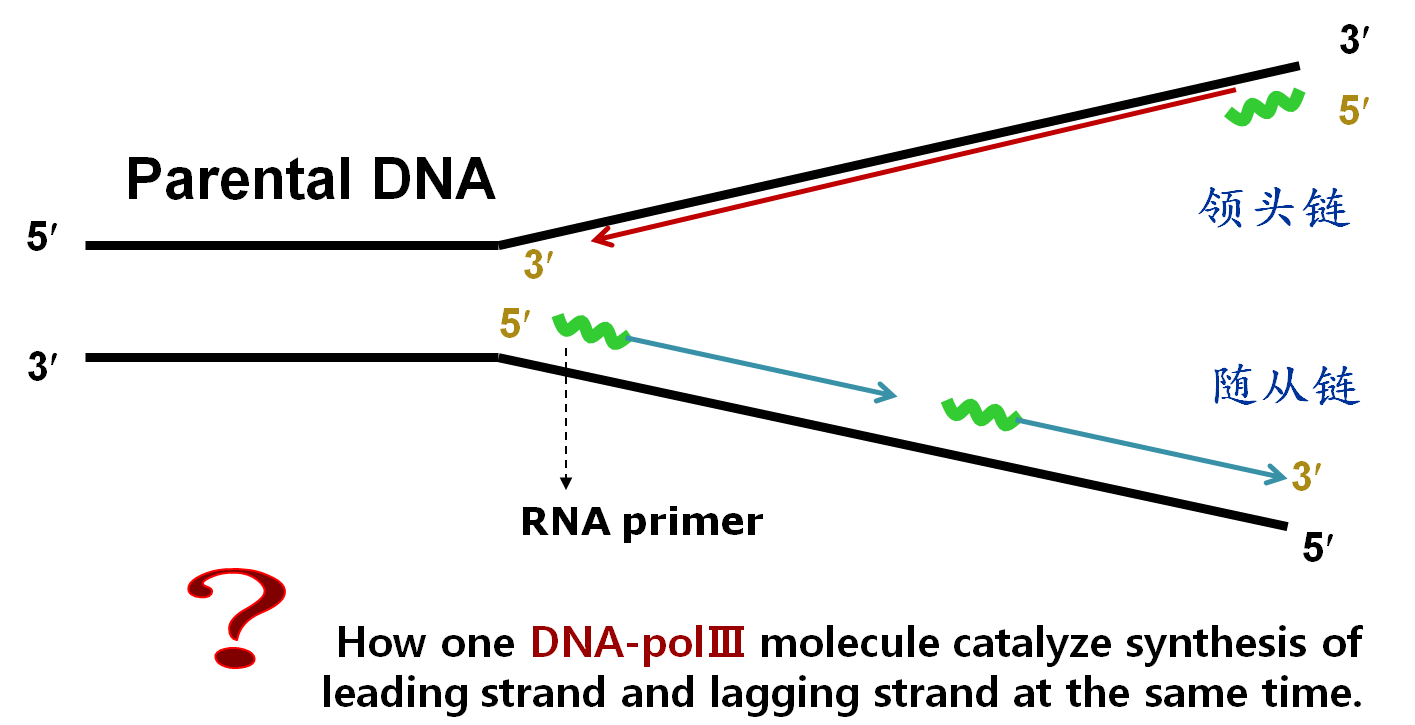

★γ complex enables DNA-primer double chain to thread through β clamp(后随链发生回折,使前导链和后随链合成方向一致).——making synthesis direction of lagging strand the sameas that of leading strand.
回折是前导链和后随链合成方向一致
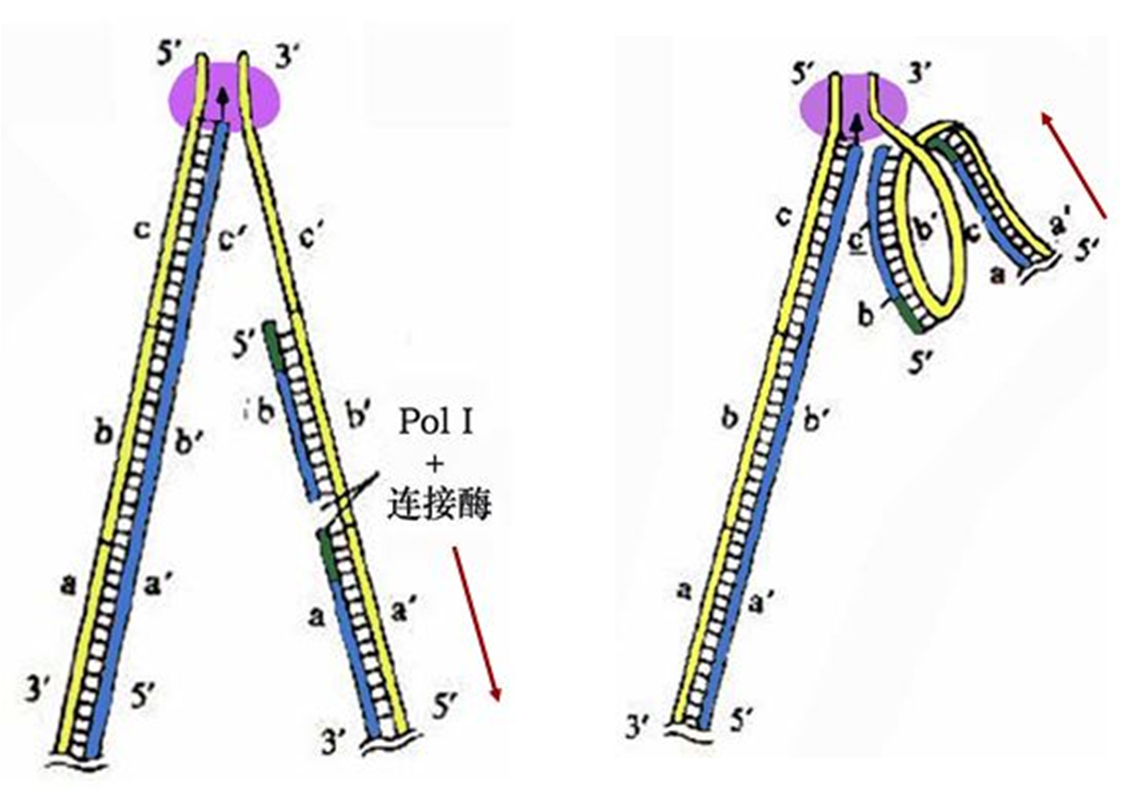
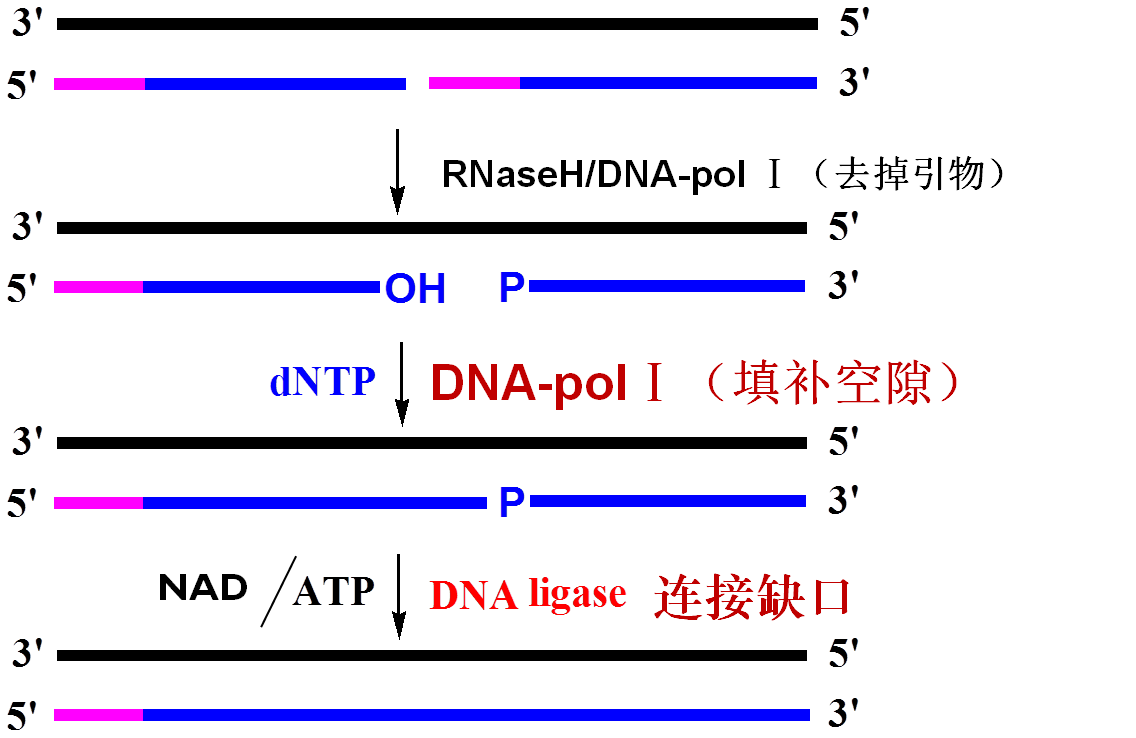 ★Primers on Okazaki fragments are digested by RNaseH or polⅠ.
★Primers on Okazaki fragments are digested by RNaseH or polⅠ.
★The gaps are filled by DNA-pol I in the 5´→3´direction.
★The nick between the 5´end of one fragment and the 3´end of the next fragment is sealed by ligase.
Termination(终止)

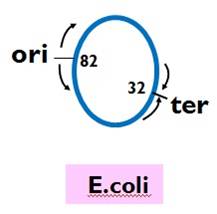
★Tus protein recognizes and binds termination site and terminates replication.
★The replication of E. coli is bidirectional from one origin, and the two replication forks must meet at one point called ter at 32.
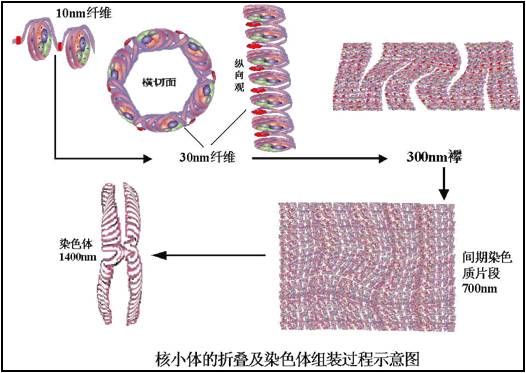
§2.2.2 Replication of eukaryotes-真核生物DNA复制

★1.Genome is much huge(3×109 bp)
★2. Genome structure is much more complicated (Nucleosome –核小体).
Elongation rate islower than that of prokaryote.

Replication process of eukaryotes 
Initiation(起始)
★1. DNA replication is closely related with cell cycle(细胞周期).
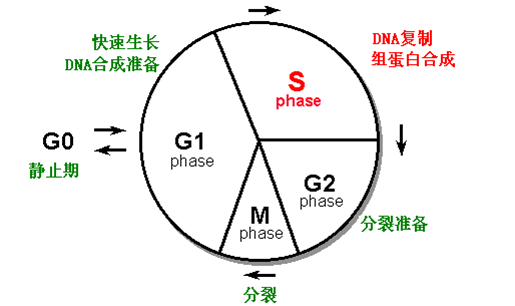
★2. Multiple origins(多复制起点) on one chromosome, and replications are activated in a sequential order rather than simultaneously.
· Ori in yeast:,ARS(autonomously replicating sequences),自主复制序列。
· ARS is recognized by origin recognition complex(ORC)-起点识别复合物
· 400 ARS in yeast genome.
· No similar sequence is found in human.
★3. Enzyme and protein factors involved in initiation.
· Helicase: Dna2(yeast), MCM(mini-chromosome maintainence protein)
· Topo
· SSB: Replication factor A(RFA)
· DNA-pol α : Sythesize primer
Elongation(延长)and Termination (终止)
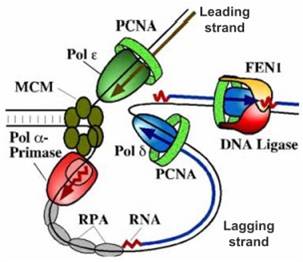
★1. Enzyme and protein factor involved in elongation.
· DNA-pol ε/δ : catalyze elongation of leading and lagging strand.
· RF-C: similar to γ complex
· PCNA(proliferating cell nuclear antigen-增殖细胞核抗原): similar to β clamp
· RNase H1 and FEN1( in human): removing primer
· DNA-pol ε/δ: gap-filing (填补空隙)
· ligase
DNA replication and nucleosome assembling(核小体装配) occur simultaneously.
Current model of DNA replication on
eukaryotic cell
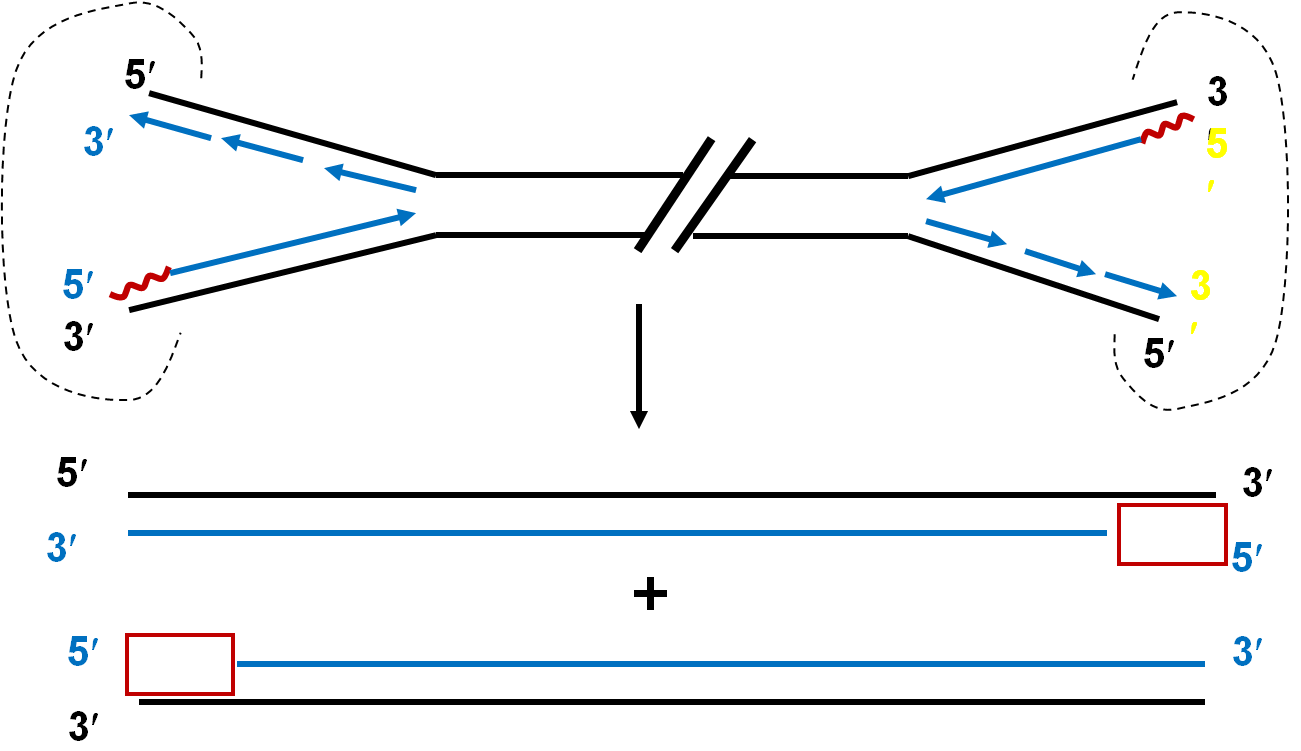
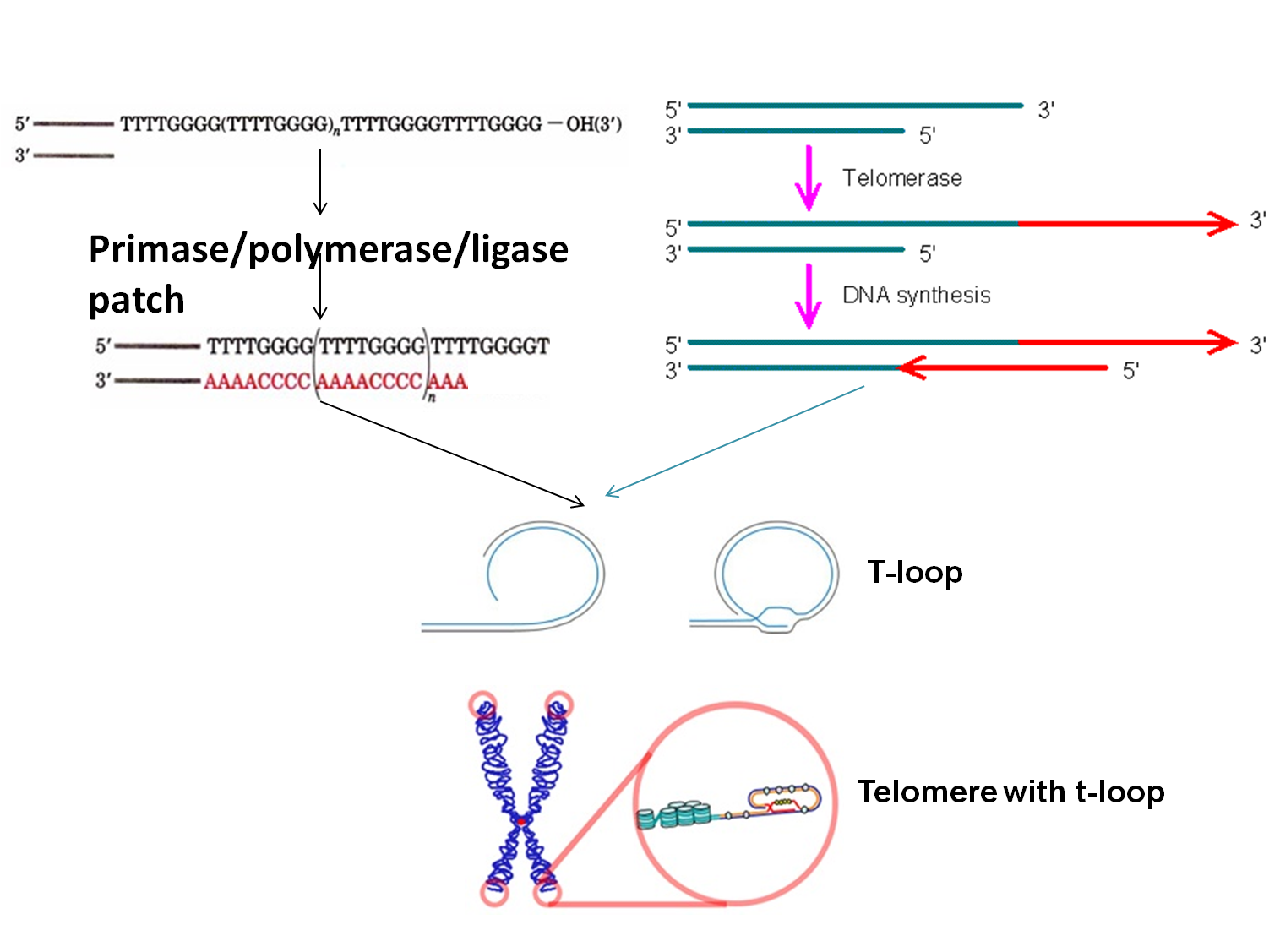
Gap filing inchromosome end
染色体末端复制的问题
§2.2.3 Telomere (端粒)

★1. Telomere
·The terminal structure of eukaryotic DNA of chromosomes is called telomere (染色体末端).
·Telomere is composed of
·The sequence of typical telomeres is rich in T and G.
·The telomere structure is crucial to keep the termini of chromosomes in the cell from becoming entangled and sticking to each other(维持染色体结构的稳定和完整).

★2. Telomerase(端粒酶)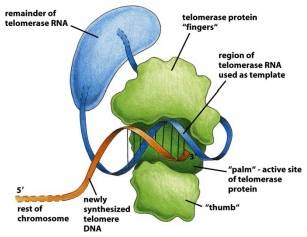
·The eukaryotic cells use telomerase to maintain the integrity of DNA telomere.
·The telomerase is composed of
![]() telomerase RNA (CA rich)
telomerase RNA (CA rich)
telomerase associationprotein
telomerase reverse transcriptase (逆转录酶)
·It is able to synthesize DNA using RNA as the template.
Inchworm model-爬行模型
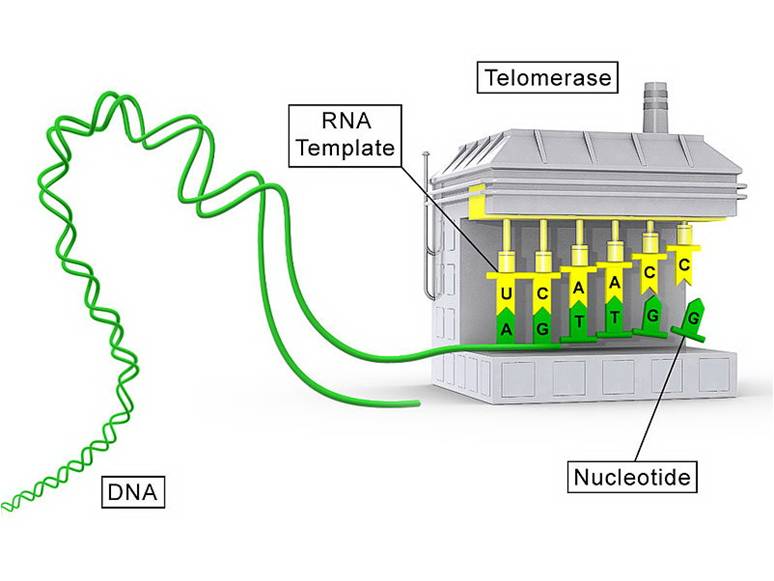
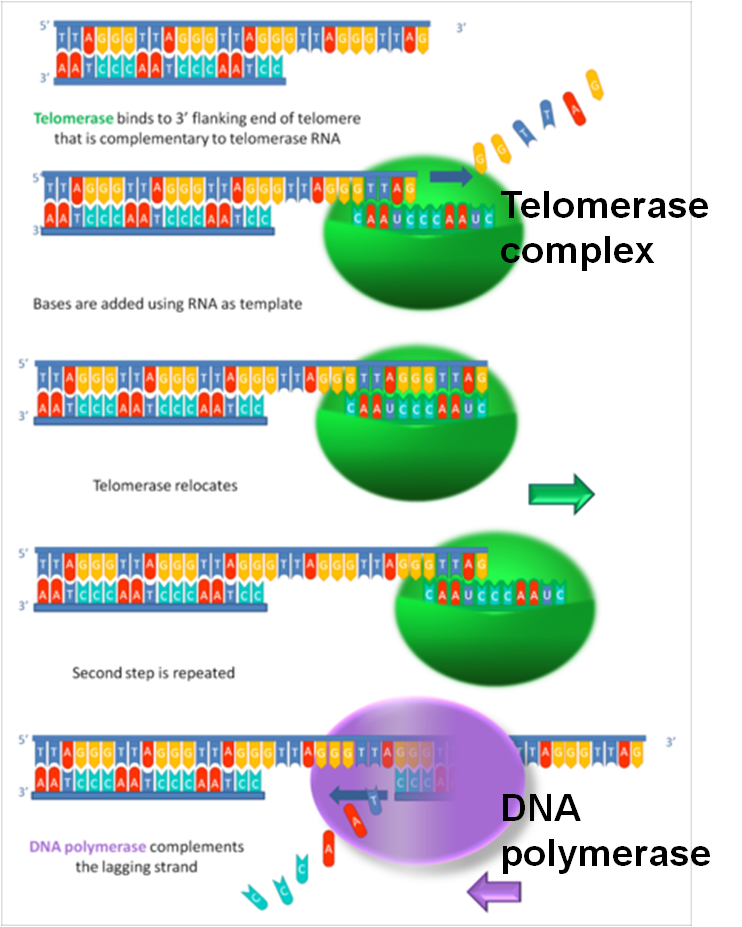 ★ 1.DNA template is extended.
★ 1.DNA template is extended.
★ 2.DNA daughter strand end is further supplemented.
Significance of Telomerase
★Telomerase may play important roles in
![]()
Cancer cell biology
Cell aging
Stem cell
§2.3 Fidelity of Replication -复制保真性

★Mechanisms to ensure the fidelity of DNA replication are:
· 1. Balanced levels of dNTPs (底物浓度,种类)
· 2. principle of base pairing is crucial to the high accuracy of the genetic information transfer. (碱基互补配对)
· 3. Base selection garanted by DNA-pol.(碱基选择)
· 4. Proofreading and mismatch correction. (校读和修复)
★1. Balanced levels of dNTPs (底物池浓度,种类)
Prereplication: dNTP pool cleansing
· 1.Balance concentration of each dNTPand total dNTP.
· 2.Avoid potentially mutagenic nucleotide.
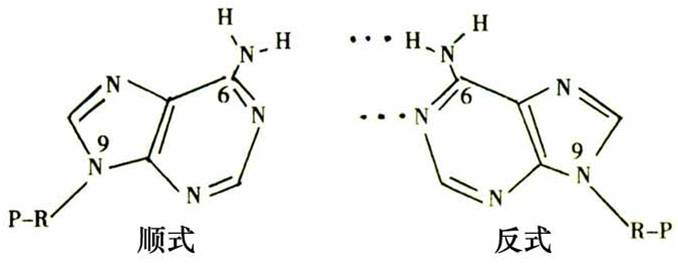
★2. 碱基互补配对
★3. 碱基选择
• DNA pol 选择正确碱基有序形成氢键.
Trans-form of A is proper for forming hydrogen bond(B-DNA).
★4. Proofreading and mismatch correction. (校读和修复-核酸外切酶功能)